Running an EWAS
There exist several approaches to run an epigenomewide association study, for example:
We have implemented our own approach DNAmArray
Collect data
The following code shows how to extract some covariates and the phenotype of interest from the SummerizedExperiments-object. Furthermore, we select a subset of the data for which the covariates and phenotype are complete, i.e. do not contain missing -values.
library(BBMRIomics)
bbmri.data(methData_Mvalues_CODAM_Freeze2_unrelated)
covariates <- c("sex", "smoking", "sentrix_position")
phenotype <- "sampling_age"
nas <- apply(as.data.frame(colData(mvalues))[,c(phenotype, covariates)], 1, anyNA)
table(nas)## nas
## FALSE TRUE
## 160 3Create design
Now we can create a design matrix appropriate for use in with limma’s lmFit-function. Both Smoking and Sentrix_Position are considered categorical variables.
## 'data.frame': 160 obs. of 4 variables:
## $ sampling_age : int 78 74 63 66 62 59 63 63 62 68 ...
## $ sex : chr "male" "male" "female" "male" ...
## $ smoking : chr "former-smoker" "former-smoker" "current smoker" "former-smoker" ...
## $ sentrix_position: chr "R05C02" "R04C01" "R05C01" "R04C01" ...design$smoking <- factor(design$smoking)
design$sentrix_position <- factor(design$sentrix_position)
str(design)## 'data.frame': 160 obs. of 4 variables:
## $ sampling_age : int 78 74 63 66 62 59 63 63 62 68 ...
## $ sex : chr "male" "male" "female" "male" ...
## $ smoking : Factor w/ 3 levels "current smoker",..: 2 2 1 2 2 1 3 2 2 2 ...
## $ sentrix_position: Factor w/ 12 levels "R01C01","R01C02",..: 10 7 9 7 1 7 2 8 1 11 ...## num [1:160, 1:16] 1 1 1 1 1 1 1 1 1 1 ...
## - attr(*, "dimnames")=List of 2
## ..$ : chr [1:160] "BIOS6DB3BAD1" "BIOSAF9A6D98" "BIOS6F87D650" "BIOS0DA7A245" ...
## ..$ : chr [1:16] "(Intercept)" "sampling_age" "sexmale" "smokingformer-smoker" ...
## - attr(*, "assign")= int [1:16] 0 1 2 3 3 4 4 4 4 4 ...
## - attr(*, "contrasts")=List of 3
## ..$ sex : chr "contr.treatment"
## ..$ smoking : chr "contr.treatment"
## ..$ sentrix_position: chr "contr.treatment"Run EWAS
Since, we had subsetted the whole SummerizedExperiments-object we can just extract the data without any further subsetting. Runnning the EWAS is now just one function-call.
Often probes on the X and Y chromosome are removed as well as cross-hybdridizing probes and probes with frequent SNPs.
mvalues <- mvalues[!(seqnames(mvalues) %in% c("chrX", "chrY")),]
data(hm450.manifest.pop.GoNL) ##From DNAmArray
hm450.manifest.pop.GoNL## GRanges object with 111799 ranges and 65 metadata columns:
## seqnames ranges strand | MASK.general.AFR MASK.snp5.AFR
## <Rle> <IRanges> <Rle> | <Rle> <Rle>
## cg13869341 chr1 15865-15866 * | TRUE FALSE
## cg14008030 chr1 18827-18828 * | TRUE FALSE
## cg12045430 chr1 29407-29408 * | TRUE FALSE
## cg20826792 chr1 29425-29426 * | TRUE FALSE
## cg00381604 chr1 29435-29436 * | TRUE FALSE
## ... ... ... ... . ... ...
## cg17939569 chrY 27009430-27009431 * | TRUE FALSE
## cg13365400 chrY 27210334-27210335 * | TRUE FALSE
## cg21106100 chrY 28555536-28555537 * | FALSE FALSE
## cg08265308 chrY 28555550-28555551 * | FALSE FALSE
## cg14273923 chrY 28555912-28555913 * | FALSE FALSE
## MASK.general.EAS MASK.snp5.EAS MASK.general.EUR MASK.snp5.EUR
## <Rle> <Rle> <Rle> <Rle>
## cg13869341 TRUE FALSE TRUE FALSE
## cg14008030 TRUE FALSE TRUE FALSE
## cg12045430 TRUE FALSE TRUE FALSE
## cg20826792 TRUE FALSE TRUE FALSE
## cg00381604 TRUE FALSE TRUE FALSE
## ... ... ... ... ...
## cg17939569 TRUE FALSE TRUE FALSE
## cg13365400 TRUE FALSE TRUE FALSE
## cg21106100 FALSE FALSE FALSE FALSE
## cg08265308 FALSE FALSE FALSE FALSE
## cg14273923 FALSE FALSE FALSE FALSE
## MASK.general.SAS MASK.snp5.SAS MASK.general.AMR MASK.snp5.AMR
## <Rle> <Rle> <Rle> <Rle>
## cg13869341 TRUE FALSE TRUE FALSE
## cg14008030 TRUE FALSE TRUE FALSE
## cg12045430 TRUE FALSE TRUE FALSE
## cg20826792 TRUE FALSE TRUE FALSE
## cg00381604 TRUE FALSE TRUE FALSE
## ... ... ... ... ...
## cg17939569 TRUE FALSE TRUE FALSE
## cg13365400 TRUE FALSE TRUE FALSE
## cg21106100 FALSE FALSE FALSE FALSE
## cg08265308 FALSE FALSE FALSE FALSE
## cg14273923 FALSE FALSE FALSE FALSE
## MASK.general.GWD MASK.snp5.GWD MASK.general.YRI MASK.snp5.YRI
## <Rle> <Rle> <Rle> <Rle>
## cg13869341 TRUE FALSE TRUE FALSE
## cg14008030 TRUE FALSE TRUE FALSE
## cg12045430 TRUE FALSE TRUE FALSE
## cg20826792 TRUE FALSE TRUE FALSE
## cg00381604 TRUE FALSE TRUE FALSE
## ... ... ... ... ...
## cg17939569 TRUE FALSE TRUE FALSE
## cg13365400 TRUE FALSE TRUE FALSE
## cg21106100 FALSE FALSE FALSE FALSE
## cg08265308 FALSE FALSE FALSE FALSE
## cg14273923 FALSE FALSE FALSE FALSE
## MASK.general.TSI MASK.snp5.TSI MASK.general.IBS MASK.snp5.IBS
## <Rle> <Rle> <Rle> <Rle>
## cg13869341 TRUE FALSE TRUE FALSE
## cg14008030 TRUE FALSE TRUE FALSE
## cg12045430 TRUE FALSE TRUE FALSE
## cg20826792 TRUE FALSE TRUE FALSE
## cg00381604 TRUE FALSE TRUE FALSE
## ... ... ... ... ...
## cg17939569 TRUE FALSE TRUE FALSE
## cg13365400 TRUE FALSE TRUE FALSE
## cg21106100 FALSE FALSE FALSE FALSE
## cg08265308 FALSE FALSE FALSE FALSE
## cg14273923 FALSE FALSE FALSE FALSE
## MASK.general.CHS MASK.snp5.CHS MASK.general.PUR MASK.snp5.PUR
## <Rle> <Rle> <Rle> <Rle>
## cg13869341 TRUE FALSE TRUE FALSE
## cg14008030 TRUE FALSE TRUE FALSE
## cg12045430 TRUE FALSE TRUE FALSE
## cg20826792 TRUE FALSE TRUE FALSE
## cg00381604 TRUE FALSE TRUE FALSE
## ... ... ... ... ...
## cg17939569 TRUE FALSE TRUE FALSE
## cg13365400 TRUE FALSE TRUE FALSE
## cg21106100 FALSE FALSE FALSE FALSE
## cg08265308 FALSE FALSE FALSE FALSE
## cg14273923 FALSE FALSE FALSE FALSE
## MASK.general.JPT MASK.snp5.JPT MASK.general.GIH MASK.snp5.GIH
## <Rle> <Rle> <Rle> <Rle>
## cg13869341 TRUE FALSE TRUE FALSE
## cg14008030 TRUE FALSE TRUE FALSE
## cg12045430 TRUE FALSE TRUE FALSE
## cg20826792 TRUE FALSE TRUE FALSE
## cg00381604 TRUE FALSE TRUE FALSE
## ... ... ... ... ...
## cg17939569 TRUE FALSE TRUE FALSE
## cg13365400 TRUE FALSE TRUE FALSE
## cg21106100 FALSE FALSE FALSE FALSE
## cg08265308 FALSE FALSE FALSE FALSE
## cg14273923 FALSE FALSE FALSE FALSE
## MASK.general.CHB MASK.snp5.CHB MASK.general.STU MASK.snp5.STU
## <Rle> <Rle> <Rle> <Rle>
## cg13869341 TRUE FALSE TRUE FALSE
## cg14008030 TRUE FALSE TRUE FALSE
## cg12045430 TRUE FALSE TRUE FALSE
## cg20826792 TRUE FALSE TRUE FALSE
## cg00381604 TRUE FALSE TRUE FALSE
## ... ... ... ... ...
## cg17939569 TRUE FALSE TRUE FALSE
## cg13365400 TRUE FALSE TRUE FALSE
## cg21106100 FALSE FALSE FALSE FALSE
## cg08265308 FALSE FALSE FALSE FALSE
## cg14273923 FALSE FALSE FALSE FALSE
## MASK.general.ITU MASK.snp5.ITU MASK.general.LWK MASK.snp5.LWK
## <Rle> <Rle> <Rle> <Rle>
## cg13869341 TRUE FALSE TRUE FALSE
## cg14008030 TRUE FALSE TRUE FALSE
## cg12045430 TRUE FALSE TRUE FALSE
## cg20826792 TRUE FALSE TRUE FALSE
## cg00381604 TRUE FALSE TRUE FALSE
## ... ... ... ... ...
## cg17939569 TRUE FALSE TRUE FALSE
## cg13365400 TRUE FALSE TRUE FALSE
## cg21106100 FALSE FALSE FALSE FALSE
## cg08265308 FALSE FALSE FALSE FALSE
## cg14273923 FALSE FALSE FALSE FALSE
## MASK.general.KHV MASK.snp5.KHV MASK.general.FIN MASK.snp5.FIN
## <Rle> <Rle> <Rle> <Rle>
## cg13869341 TRUE FALSE TRUE FALSE
## cg14008030 TRUE FALSE TRUE FALSE
## cg12045430 TRUE FALSE TRUE FALSE
## cg20826792 TRUE FALSE TRUE FALSE
## cg00381604 TRUE FALSE TRUE FALSE
## ... ... ... ... ...
## cg17939569 TRUE FALSE TRUE FALSE
## cg13365400 TRUE FALSE TRUE FALSE
## cg21106100 FALSE FALSE FALSE FALSE
## cg08265308 FALSE FALSE FALSE FALSE
## cg14273923 FALSE FALSE FALSE FALSE
## MASK.general.ESN MASK.snp5.ESN MASK.general.CEU MASK.snp5.CEU
## <Rle> <Rle> <Rle> <Rle>
## cg13869341 TRUE FALSE TRUE FALSE
## cg14008030 TRUE FALSE TRUE FALSE
## cg12045430 TRUE FALSE TRUE FALSE
## cg20826792 TRUE FALSE TRUE FALSE
## cg00381604 TRUE FALSE TRUE FALSE
## ... ... ... ... ...
## cg17939569 TRUE FALSE TRUE FALSE
## cg13365400 TRUE FALSE TRUE FALSE
## cg21106100 FALSE FALSE FALSE FALSE
## cg08265308 FALSE FALSE FALSE FALSE
## cg14273923 FALSE FALSE FALSE FALSE
## MASK.general.PJL MASK.snp5.PJL MASK.general.ACB MASK.snp5.ACB
## <Rle> <Rle> <Rle> <Rle>
## cg13869341 TRUE FALSE TRUE FALSE
## cg14008030 TRUE FALSE TRUE FALSE
## cg12045430 TRUE FALSE TRUE FALSE
## cg20826792 TRUE FALSE TRUE FALSE
## cg00381604 TRUE FALSE TRUE FALSE
## ... ... ... ... ...
## cg17939569 TRUE FALSE TRUE FALSE
## cg13365400 TRUE FALSE TRUE FALSE
## cg21106100 FALSE FALSE FALSE FALSE
## cg08265308 FALSE FALSE FALSE FALSE
## cg14273923 FALSE FALSE FALSE FALSE
## MASK.general.CLM MASK.snp5.CLM MASK.general.CDX MASK.snp5.CDX
## <Rle> <Rle> <Rle> <Rle>
## cg13869341 TRUE FALSE TRUE FALSE
## cg14008030 TRUE FALSE TRUE FALSE
## cg12045430 TRUE FALSE TRUE FALSE
## cg20826792 TRUE FALSE TRUE FALSE
## cg00381604 TRUE FALSE TRUE FALSE
## ... ... ... ... ...
## cg17939569 TRUE FALSE TRUE FALSE
## cg13365400 TRUE FALSE TRUE FALSE
## cg21106100 FALSE FALSE FALSE FALSE
## cg08265308 FALSE FALSE FALSE FALSE
## cg14273923 FALSE FALSE FALSE FALSE
## MASK.general.GBR MASK.snp5.GBR MASK.general.BEB MASK.snp5.BEB
## <Rle> <Rle> <Rle> <Rle>
## cg13869341 TRUE FALSE TRUE FALSE
## cg14008030 TRUE FALSE TRUE FALSE
## cg12045430 TRUE FALSE TRUE FALSE
## cg20826792 TRUE FALSE TRUE FALSE
## cg00381604 TRUE FALSE TRUE FALSE
## ... ... ... ... ...
## cg17939569 TRUE FALSE TRUE FALSE
## cg13365400 TRUE FALSE TRUE FALSE
## cg21106100 FALSE FALSE FALSE FALSE
## cg08265308 FALSE FALSE FALSE FALSE
## cg14273923 FALSE FALSE FALSE FALSE
## MASK.general.PEL MASK.snp5.PEL MASK.general.MSL MASK.snp5.MSL
## <Rle> <Rle> <Rle> <Rle>
## cg13869341 TRUE FALSE TRUE FALSE
## cg14008030 TRUE FALSE TRUE FALSE
## cg12045430 TRUE FALSE TRUE FALSE
## cg20826792 TRUE FALSE TRUE FALSE
## cg00381604 TRUE FALSE TRUE FALSE
## ... ... ... ... ...
## cg17939569 TRUE FALSE TRUE FALSE
## cg13365400 TRUE FALSE TRUE FALSE
## cg21106100 FALSE FALSE FALSE FALSE
## cg08265308 FALSE FALSE FALSE FALSE
## cg14273923 FALSE FALSE FALSE FALSE
## MASK.general.MXL MASK.snp5.MXL MASK.general.ASW MASK.snp5.ASW
## <Rle> <Rle> <Rle> <Rle>
## cg13869341 TRUE FALSE TRUE FALSE
## cg14008030 TRUE FALSE TRUE FALSE
## cg12045430 TRUE FALSE TRUE FALSE
## cg20826792 TRUE FALSE TRUE FALSE
## cg00381604 TRUE FALSE TRUE FALSE
## ... ... ... ... ...
## cg17939569 TRUE FALSE TRUE FALSE
## cg13365400 TRUE FALSE TRUE FALSE
## cg21106100 FALSE FALSE FALSE FALSE
## cg08265308 FALSE FALSE FALSE FALSE
## cg14273923 FALSE FALSE FALSE FALSE
## MASK.snp5.GoNL MASK.general.GoNL
## <Rle> <Rle>
## cg13869341 FALSE TRUE
## cg14008030 FALSE TRUE
## cg12045430 FALSE TRUE
## cg20826792 FALSE TRUE
## cg00381604 FALSE TRUE
## ... ... ...
## cg17939569 <NA> <NA>
## cg13365400 <NA> <NA>
## cg21106100 <NA> <NA>
## cg08265308 <NA> <NA>
## cg14273923 <NA> <NA>
## MASK.typeINextBaseSwitchandINDEL.GoNL
## <Rle>
## cg13869341 FALSE
## cg14008030 FALSE
## cg12045430 FALSE
## cg20826792 FALSE
## cg00381604 FALSE
## ... ...
## cg17939569 <NA>
## cg13365400 <NA>
## cg21106100 <NA>
## cg08265308 <NA>
## cg14273923 <NA>
## -------
## seqinfo: 25 sequences from an unspecified genome; no seqlengthsInspect results
Since, we have over 100 samples we ignore the usual steps of obtaining regularized test-statistics using the function eBayes. The ordinary t-statistics can be extracted as follows (as described in the vignette of limma):
tstat <- fit$coef/fit$stdev.unscaled/fit$sigma
pval <- 2 * pnorm(-abs(tstat[, 2]))
padj <- p.adjust(sort(pval, decreasing = FALSE), method = "bonf")
head(padj[padj < 0.05])## cg16867657 cg22454769 cg06639320 cg08097417 cg04875128 cg21572722
## 1.149202e-23 2.978821e-09 5.589675e-08 7.011138e-08 3.379335e-07 8.740024e-07After preforming multiple-testing correction using Bonferroni’s method we can inspect the results.
gp <- ggplot(data.frame(pval=pval), aes(sample=-log10(pval)))
gp <- gp + stat_qq(distribution=stats::qexp, dparams=list(rate=1/log10(exp(1))))
gp <- gp + xlab(expression(paste("Expected -log"[10], plain(P))))
gp <- gp + ylab(expression(paste("Observed -log"[10], plain(P))))
gp <- gp + geom_abline(slope=1, intercept=0)
gp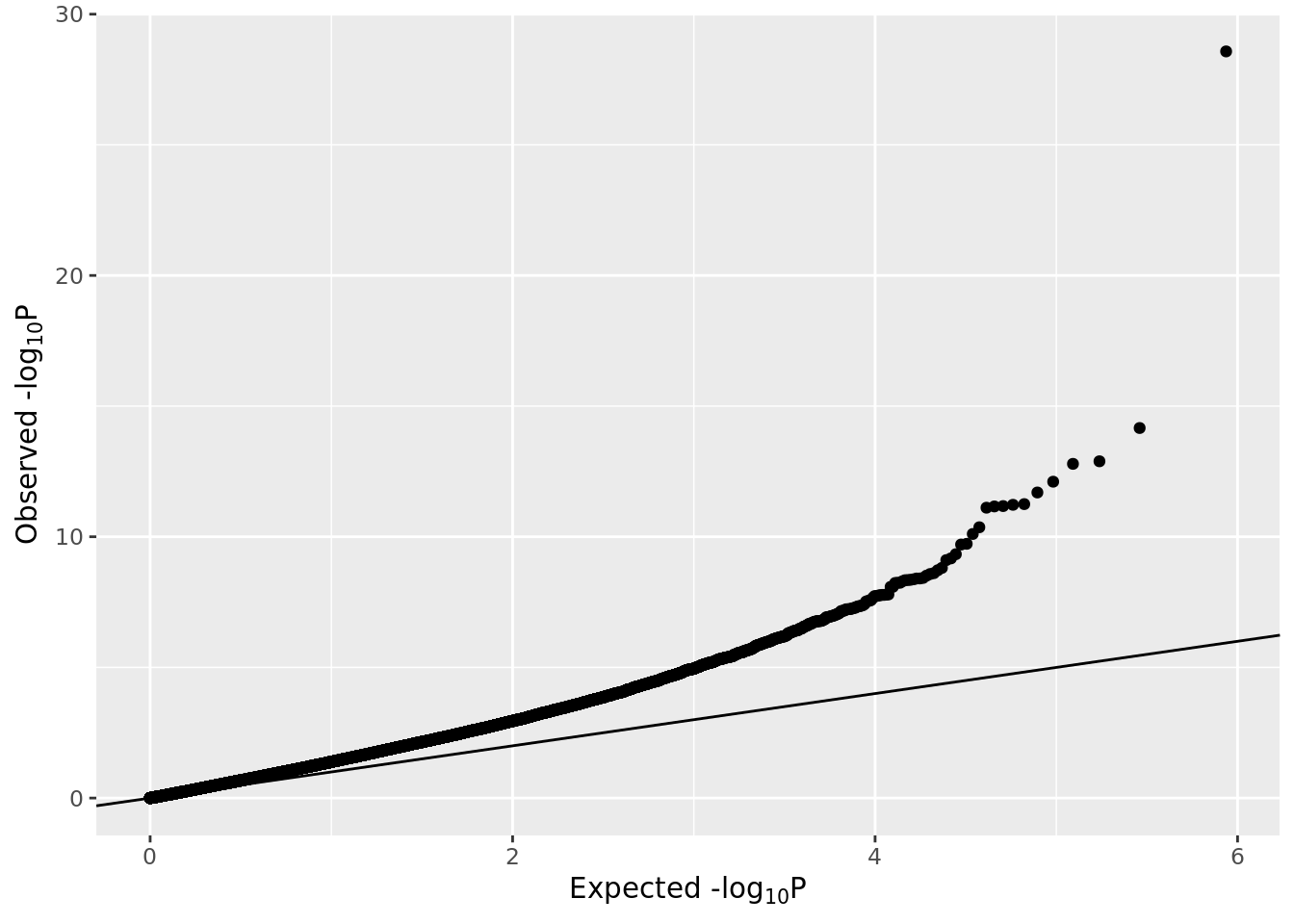
Or make an manhattan plot:
## GRanges object with 432376 ranges and 10 metadata columns:
## seqnames ranges strand | addressA addressB
## <Rle> <IRanges> <Rle> | <character> <character>
## cg00000957 chr1 5937253-5937254 * | 65648367 36743439
## cg00001349 chr1 166958439-166958440 * | 11722421 53758324
## cg00001583 chr1 200011786-200011787 * | 55630379 24638471
## cg00002028 chr1 20960010-20960011 * | 56738397 74621430
## cg00002719 chr1 169396706-169396707 * | 52741444 27736432
## ... ... ... ... . ... ...
## ch.22.44116734F chr22 45738070 * | 61782438
## ch.22.909671F chr22 46114168 * | 47797398
## ch.22.46830341F chr22 48451677 * | 29618504
## ch.22.1008279F chr22 48731367 * | 49664383
## ch.22.47579720R chr22 49193714 * | 53733426
## channel platform percentGC sourceSeq probeType
## <Rle> <Rle> <numeric> <DNAStringSet> <Rle>
## cg00000957 Grn HM450 0.7 ATGCTACTGA...ACGTCAGCCG cg
## cg00001349 Grn HM450 0.62 CAAGGCGGCA...TGTTCCCCCG cg
## cg00001583 Red HM450 0.7 CGGCGAAACC...TACCTCCTGC cg
## cg00002028 Red HM450 0.74 CGCCGCTGCC...CAAACTTGGG cg
## cg00002719 Red HM450 0.56 CGAGAGCGAC...CAGACCACCG cg
## ... ... ... ... ... ...
## ch.22.44116734F Both HM450 0.44 CAAGCATAGA...TACAGCCCAT ch
## ch.22.909671F Both HM450 0.34 CAGCAAATCA...GTAAGTGGTG ch
## ch.22.46830341F Both HM450 0.46 CAGCATCACA...TCCATTTTTC ch
## ch.22.1008279F Both HM450 0.56 CAAGACTCAT...GACTGTAGGG ch
## ch.22.47579720R Both HM450 0.6 CAGGCAAGGG...CTGGAGAGAG ch
## probeStart probeEnd probeTarget
## <character> <character> <numeric>
## cg00000957 5937253 5937302 5937253
## cg00001349 166958391 166958440 166958439
## cg00001583 200011738 200011787 200011786
## cg00002028 20959962 20960011 20960010
## cg00002719 169396706 169396755 169396706
## ... ... ... ...
## ch.22.44116734F 45738070 45738119 45738070
## ch.22.909671F 46114168 46114217 46114168
## ch.22.46830341F 48451677 48451726 48451677
## ch.22.1008279F 48731367 48731416 48731367
## ch.22.47579720R 49193714 49193763 49193714
## -------
## seqinfo: 24 sequences from hg19 genome## cg00000957 cg00001349 cg00001583 cg00002028 cg00002719 cg00002837
## 0.8635208605 0.2600199848 0.0001467086 0.0292046930 0.0704876977 0.0001973381## ch.22.43177094F ch.22.44116734F ch.22.909671F ch.22.46830341F ch.22.1008279F
## 0.3255134 0.9535643 0.5900916 0.8197218 0.8665827
## ch.22.47579720R
## 0.4525095## GRanges object with 432376 ranges and 11 metadata columns:
## seqnames ranges strand | addressA addressB
## <Rle> <IRanges> <Rle> | <character> <character>
## cg00000957 chr1 5937253-5937254 * | 65648367 36743439
## cg00001349 chr1 166958439-166958440 * | 11722421 53758324
## cg00001583 chr1 200011786-200011787 * | 55630379 24638471
## cg00002028 chr1 20960010-20960011 * | 56738397 74621430
## cg00002719 chr1 169396706-169396707 * | 52741444 27736432
## ... ... ... ... . ... ...
## ch.22.44116734F chr22 45738070 * | 61782438
## ch.22.909671F chr22 46114168 * | 47797398
## ch.22.46830341F chr22 48451677 * | 29618504
## ch.22.1008279F chr22 48731367 * | 49664383
## ch.22.47579720R chr22 49193714 * | 53733426
## channel platform percentGC sourceSeq probeType
## <Rle> <Rle> <numeric> <DNAStringSet> <Rle>
## cg00000957 Grn HM450 0.7 ATGCTACTGA...ACGTCAGCCG cg
## cg00001349 Grn HM450 0.62 CAAGGCGGCA...TGTTCCCCCG cg
## cg00001583 Red HM450 0.7 CGGCGAAACC...TACCTCCTGC cg
## cg00002028 Red HM450 0.74 CGCCGCTGCC...CAAACTTGGG cg
## cg00002719 Red HM450 0.56 CGAGAGCGAC...CAGACCACCG cg
## ... ... ... ... ... ...
## ch.22.44116734F Both HM450 0.44 CAAGCATAGA...TACAGCCCAT ch
## ch.22.909671F Both HM450 0.34 CAGCAAATCA...GTAAGTGGTG ch
## ch.22.46830341F Both HM450 0.46 CAGCATCACA...TCCATTTTTC ch
## ch.22.1008279F Both HM450 0.56 CAAGACTCAT...GACTGTAGGG ch
## ch.22.47579720R Both HM450 0.6 CAGGCAAGGG...CTGGAGAGAG ch
## probeStart probeEnd probeTarget pval
## <character> <character> <numeric> <numeric>
## cg00000957 5937253 5937302 5937253 0.863520860477633
## cg00001349 166958391 166958440 166958439 0.260019984842207
## cg00001583 200011738 200011787 200011786 0.000146708565109417
## cg00002028 20959962 20960011 20960010 0.029204693044866
## cg00002719 169396706 169396755 169396706 0.0704876977162638
## ... ... ... ... ...
## ch.22.44116734F 45738070 45738119 45738070 0.953564271207452
## ch.22.909671F 46114168 46114217 46114168 0.590091601585492
## ch.22.46830341F 48451677 48451726 48451677 0.819721782308882
## ch.22.1008279F 48731367 48731416 48731367 0.866582690269309
## ch.22.47579720R 49193714 49193763 49193714 0.452509478635683
## -------
## seqinfo: 24 sequences from hg19 genome## GRanges object with 432376 ranges and 11 metadata columns:
## seqnames ranges strand | addressA addressB
## <Rle> <IRanges> <Rle> | <character> <character>
## cg00000957 1 5937253-5937254 * | 65648367 36743439
## cg00001349 1 166958439-166958440 * | 11722421 53758324
## cg00001583 1 200011786-200011787 * | 55630379 24638471
## cg00002028 1 20960010-20960011 * | 56738397 74621430
## cg00002719 1 169396706-169396707 * | 52741444 27736432
## ... ... ... ... . ... ...
## ch.22.44116734F 22 45738070 * | 61782438
## ch.22.909671F 22 46114168 * | 47797398
## ch.22.46830341F 22 48451677 * | 29618504
## ch.22.1008279F 22 48731367 * | 49664383
## ch.22.47579720R 22 49193714 * | 53733426
## channel platform percentGC sourceSeq probeType
## <Rle> <Rle> <numeric> <DNAStringSet> <Rle>
## cg00000957 Grn HM450 0.7 ATGCTACTGA...ACGTCAGCCG cg
## cg00001349 Grn HM450 0.62 CAAGGCGGCA...TGTTCCCCCG cg
## cg00001583 Red HM450 0.7 CGGCGAAACC...TACCTCCTGC cg
## cg00002028 Red HM450 0.74 CGCCGCTGCC...CAAACTTGGG cg
## cg00002719 Red HM450 0.56 CGAGAGCGAC...CAGACCACCG cg
## ... ... ... ... ... ...
## ch.22.44116734F Both HM450 0.44 CAAGCATAGA...TACAGCCCAT ch
## ch.22.909671F Both HM450 0.34 CAGCAAATCA...GTAAGTGGTG ch
## ch.22.46830341F Both HM450 0.46 CAGCATCACA...TCCATTTTTC ch
## ch.22.1008279F Both HM450 0.56 CAAGACTCAT...GACTGTAGGG ch
## ch.22.47579720R Both HM450 0.6 CAGGCAAGGG...CTGGAGAGAG ch
## probeStart probeEnd probeTarget pval
## <character> <character> <numeric> <numeric>
## cg00000957 5937253 5937302 5937253 0.863520860477633
## cg00001349 166958391 166958440 166958439 0.260019984842207
## cg00001583 200011738 200011787 200011786 0.000146708565109417
## cg00002028 20959962 20960011 20960010 0.029204693044866
## cg00002719 169396706 169396755 169396706 0.0704876977162638
## ... ... ... ... ...
## ch.22.44116734F 45738070 45738119 45738070 0.953564271207452
## ch.22.909671F 46114168 46114217 46114168 0.590091601585492
## ch.22.46830341F 48451677 48451726 48451677 0.819721782308882
## ch.22.1008279F 48731367 48731416 48731367 0.866582690269309
## ch.22.47579720R 49193714 49193763 49193714 0.452509478635683
## -------
## seqinfo: 24 sequences from hg19 genomeseqlevels(rData) <- gsub("X", 23, seqlevels(rData))
seqlevels(rData) <- gsub("Y", 24, seqlevels(rData))
pdata <- as.data.frame(rData)
pdata$seqnames <- as.integer(pdata$seqnames)
library(qqman)
manhattan(pdata, chr="seqnames", bp="start", p="pval")## Warning in manhattan(pdata, chr = "seqnames", bp = "start", p = "pval"): No SNP
## column found. OK unless you're trying to highlight.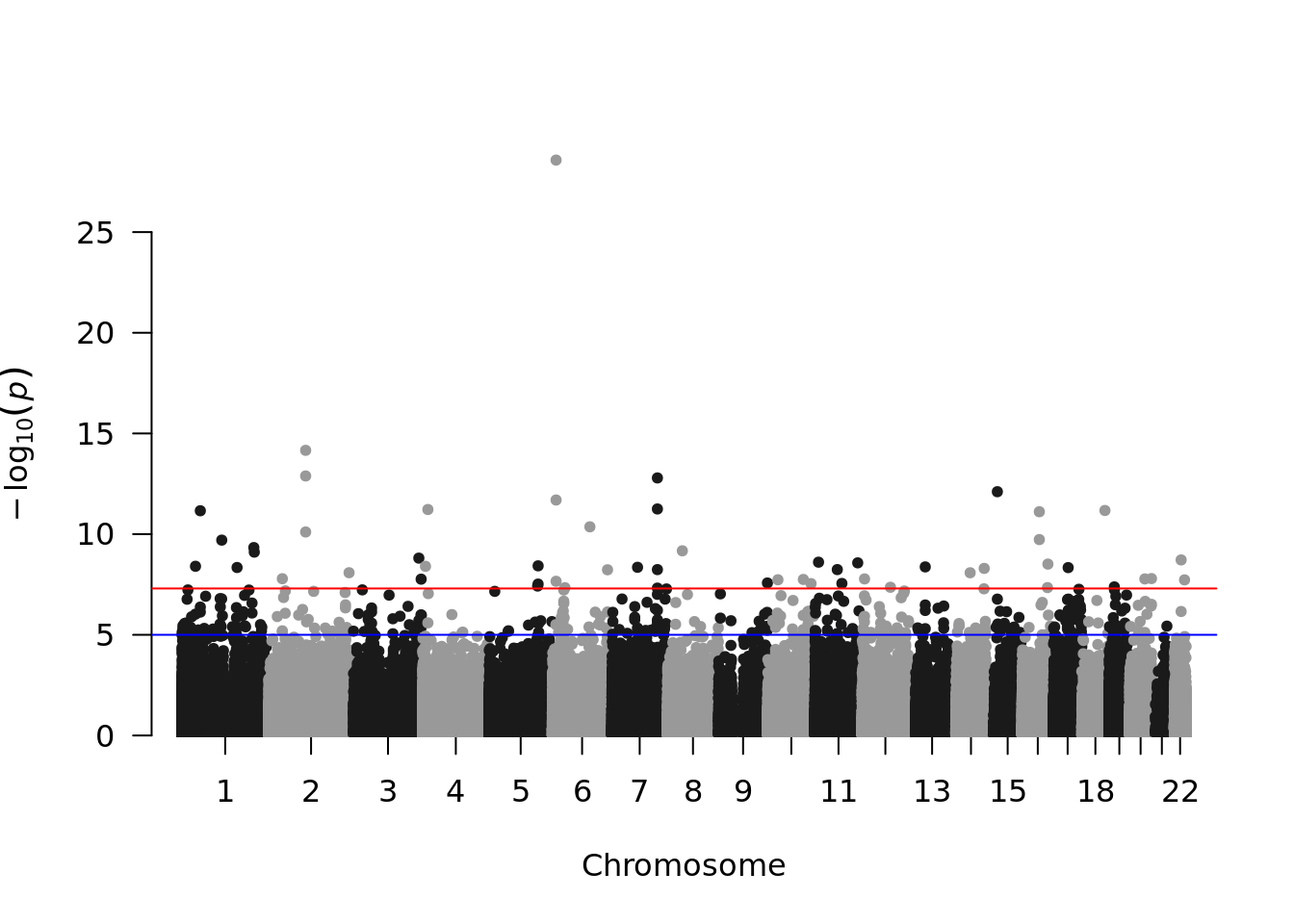
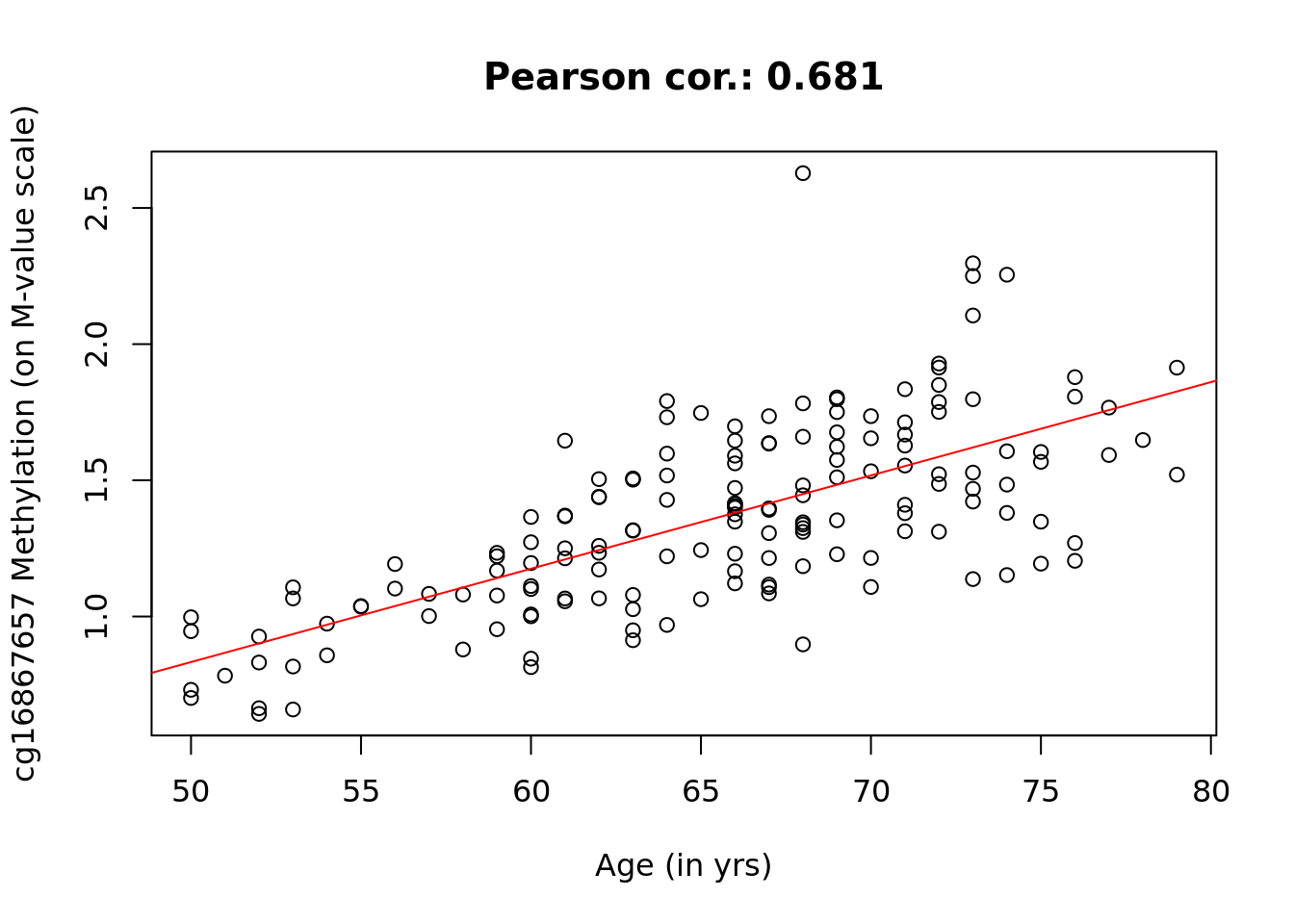
The famous CpG near the ELOVL2 gene.
top <- which.min(rData$pval)
y <- data[top, ]
x <- design[, 2]
plot(x, y, main = paste("Pearson cor.:", signif(cor(x, y), 3)), xlab = "Age (in yrs)",
ylab = paste(rownames(data)[top], "Methylation (on M-value scale)"))
abline(lm(y ~ x), col = 2)